RNA model
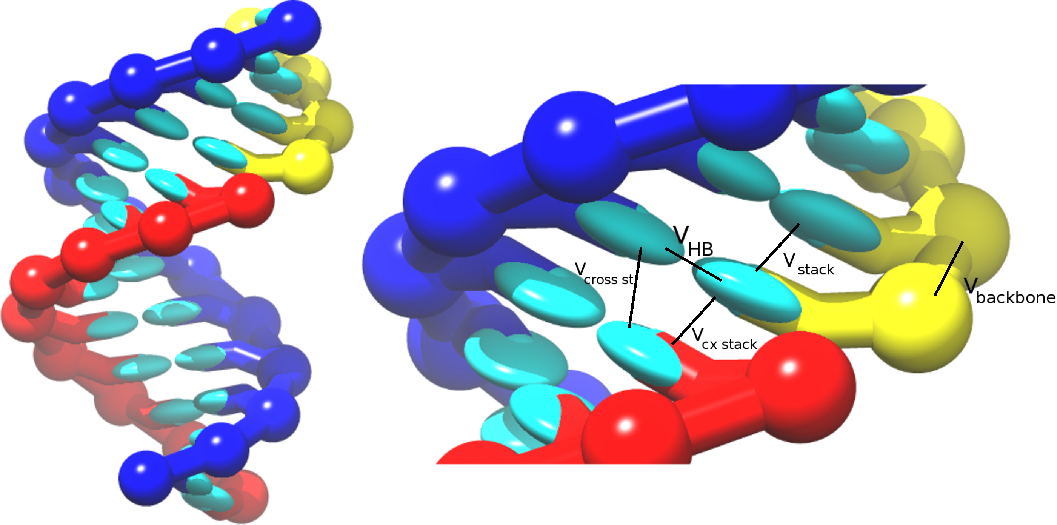
The RNA model, oxRNA, treats each RNA nucleotide as a single rigid body with multiple interaction sites, following the coarse-graining apporach adopted for the DNA model. The nucleotides interact with pairwise interaction potentials, which are schematically illustrated below: [[Image::Image_duplex_combined_annotated.png]]
- Backbone connectivity V_(b.b.),
- Excluded volume ,
- Hydrogen bonding,
- Nearest-neighbour stacking,
- Cross-stacking between base-pair steps in a duplex,
- Coaxial stacking.
which are schematically illustrated in the picture:
Simulation units
The code uses dimensionless energy, mass, length and timescales for convenience. The relationship between simulation units (SU) and SI units is given below.
| Simulation unit | Physical unit |
|---|---|
| 1 unit of length | 8.518x10 m |
| 1 unit of energy | 4.142x10 J |
| 1 unit of temperature | 3000 K |
| 1 unit of force | 4.863x10 N |
| 1 unit of mass | 1.66x10 kg |
| 1 unit of time | 1.71x10 s |
The model and its performance is discussed in detail in the following references (the thesis provides the most complete analysis):
T. E. Ouldridge, D.Phil. Thesis, University of Oxford, 2011. Coarse-grained modelling of DNA and DNA self-assembly
T. E. Ouldridge, A. A. Louis and J. P. K. Doye, J. Chem. Phys, 134, 085101 (2011) Structural, mechanical and thermodynamic properties of a coarse-grained DNA model (arXiv)
P. Šulc, F. Romano, T. E. Ouldridge, L. Rovigatti, J. P. K. Doye, A. A. Louis, J. Chem. Phys. 137, 135101 (2012) Sequence-dependent thermodynamics of a coarse-grained DNA model (arxiv)