Cadnano: Difference between revisions
No edit summary |
|||
| Line 39: | Line 39: | ||
Note that using this interaction to relax structures can be tricky and the difficulty varies depending on the design. It may be necessary to use a lot of trial and error, to modify the cadnano file, and to use external forces to guide the origami towards the correct structure. | Note that using this interaction to relax structures can be tricky and the difficulty varies depending on the design. It may be necessary to use a lot of trial and error, to modify the cadnano file, and to use external forces to guide the origami towards the correct structure. | ||
== Updated relaxation procedure == | |||
The latest recommended approach to relaxing origamis and other structures can be found here: ${oxDNA}/EXAMPLES/RELAX_INITIAL_CONFIGURATION/ directory. | |||
== Choosing the sequence == | == Choosing the sequence == | ||
Revision as of 10:10, 24 October 2018
Cadnano is a tool for designing DNA origami structures. oxDNA includes an interface that allows origami designs generated in cadnano to be used as starting configurations for simulation. Cadnano can also be used in this way to make non-origami structures such as DNA tiles for use in the model.
Example: DX tile

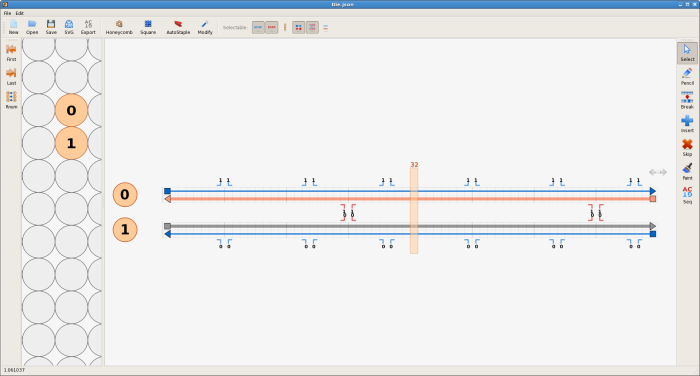
The use of cadnano for the purpose of creating starting configurations will be illustrated using a tile similar to the DX tile. In this case cadnano 2 will be used to design the tile, although the original cadnano operates in a similar way and is fully compatible with the interface to oxDNA. Note that, while using cadnano, any unwanted actions can be undone with ctrl+z (this feature is new in cadnano 2).
First click the blue "add new square lattice" button on the toolbar - a new square lattice will be created in the lattice view. The circles represent a cross-sectional view of potential DNA double helices on the square lattice. Create two empty virtual double helices in the path view by clicking on any two adjacent circles, one on top of the other. Next, using the pencil tool from the toolbar on the right, fill in all four rows in the path view by dragging from one end of each row to the other. Each double helix has two rows, and each row represents one of the strands of a double helix.
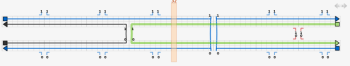
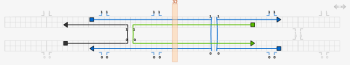
The next step is to add the crossovers between the double helices. Using origami terminology, the thin blue lines represent the scaffold strands, while the thicker lines of different colour represent the staple strands. Make a crossover between the staple strands near the middle of the design, by first clicking on one of the staple strands near its middle and then clicking one of the numbers near the middle next to that staple strand. Also click the number just to the left or right of the crossover you just made, to make a second crossover. Do the same for the the scaffold strands - the crossovers can go either to the left or the right of the staple crossovers, not at the closest place to the staple crossovers, but the next closest place.
The tile is almost complete; the final step is to correct the lengths of the strands. Switch to the select tool and drag the ends of the strands to change their lengths. The two strands that are complementary to a double crossover should extend 8 bases past that crossover. The other two strands should extend 5 bases past those two strands. The yellow bar can help with tasks like this; drag it to any column to see the index of that column. If extra space is required, the virtual helices can be extended in either direction by clicking on the arrows at the top right of the topmost virtual helix in the path view.
The tile design is now finished and ready to be used as a starting configuration for simulation after some processing. The design can be found at ${oxDNA}/EXAMPLES/CADNANO_INTERFACE/TILE/tile.json.
Using Cadnano Designs as Starting Configurations for oxDNA
The script candnano_interface.py is used to generate an oxDNA configuration and topology file from a cadnano design. In addition it creates some files containing information about the origami that can be useful later for analysing its trajectory. Its usage is
python cadnano_interface.py <cadnano_file> <design_type> [box_size]
The design type must be either sq or he, corresponding to either a square or honeycomb lattice - typically a 2D origami is on a square lattice, while a 3D origami is on a honeycomb lattice. The box size option allows the user to specify a simulation box size different to the default value of two times (in linear dimension) the largest dimension of the cadnano design.
When the configuration and topology files are first created, they cannot be used in an ordinary oxDNA simulation until they have been relaxed. This is achieved using an oxDNA MD simulation with a very low temperature and a very strongly coupled thermostat. An example input file for this simulation is found at ${oxDNA}/EXAMPLES/CADNANO_INTERFACE/input_relax. The process is very fast as the simulation need only run for around 100 steps.
For the tile discussed above, the configuration files before and after relaxation, as well as the topology file, can be found in ${oxDNA}/EXAMPLES/CADNANO_INTERFACE/TILE/.
Designs with forced crossovers
Forced crossovers, that is crossovers made by the pencil tool instead of using the default cadnano default positions, will probably create a structure that cannot be relaxed using the input_relax method detailed in the previous section. There is now a way to relax these structures too, by using an alternative interaction_type with a custom interaction term between neighbours on a strand. An example and more detailed description are provided in the ${oxDNA}/EXAMPLES/RELAX_INITIAL_CONFIGURATION/ directory.
Note that using this interaction to relax structures can be tricky and the difficulty varies depending on the design. It may be necessary to use a lot of trial and error, to modify the cadnano file, and to use external forces to guide the origami towards the correct structure.
Updated relaxation procedure
The latest recommended approach to relaxing origamis and other structures can be found here: ${oxDNA}/EXAMPLES/RELAX_INITIAL_CONFIGURATION/ directory.
Choosing the sequence
By default, a scaffold strand with a random sequence is generated. It is possible to set the sequence instead. The script looks for a file containing the sequence called "caca.sqs". That file should contain a letter (A,T,G,C or R for random) designating the sequence of the scaffold strand, for every pair of squares (which denote a base pair) in the cadnano design. This includes squares that do not contain a strand. So the sequence file should be an M by N grid of letters where M is the length of the helices and N is the number of helices in the cadnano design (including entirely unoccupied rows). Note that there is no need to include skips in the sequence file.
Limitations
- For very large cadnano designs, or for designs of small origamis but with very long unused sections of virtual double helices, the script requires a very large amount of memory which may cause problems.
- A large number of insertions or deletions on a small section of dna (where a section is defined as the bases between nodes, a node being a crossover or the end of a strand) may create a structure that cannot be relaxed.
- Very large structures with very frequent crossovers may create a structure that cannot be relaxed.